 DR_MICROBE / ISTOCK / GETTY IMAGES PLUS
DR_MICROBE / ISTOCK / GETTY IMAGES PLUS
The Pathogenicity of Porphyromonas Gingivalis
This Gram-negative anaerobe plays a significant role in inflammation and disease.
Part 1 of a two-part series. This article is Part 1 of a two-part series on Porphyromonas gingivalis. Part 2 will explore its systemic impacts.
This course was published in the October 2022 issue and expires October 2025. The authors have no commercial conflicts of interest to disclose. This 2 credit hour self-study activity is electronically mediated.
AGD Subject Code: 490
EDUCATIONAL OBJECTIVES
After reading this course, the participant should be able to:
- Describe the characteristics of Porphyromonas gingivalis.
- Discuss P. gingivalis’ virulence factors.
- Identify the roles of the T9SS system and peptidylarginine deiminase.
Acknowledged as one of the most important bacteria in the etiology and pathogenesis of periodontal diseases, Porphyromonas gingivalis continues to be extensively studied, not only in periodontal diseases, but also in many systemic diseases based on its ability to infect distant tissues and organs.1–5 It is a highly destructive oral pathogen, and, together with Treponema denticola and Tannerella forsythia, it comprises the “red complex” bacteria associated with severe periodontitis.6
Although able to initiate periodontal diseases even at low levels in biofilm, P. gingivalis nevertheless requires the presence of other bacteria to cause disease.7 It also promotes the development of “pathobiont” species, or bacteria that become pathogenic in the presence of P. gingivalis. This Gram-negative anaerobe accomplishes this by altering the entire microbial community and triggering bacterial dysbiosis.8,9
The bacterium’s unique features offer new insights into the development and progression of periodontal diseases and the oral-systemic link.4,5,8,10,11 While the connection between oral and systemic health is not a new discovery, the process by which periodontal pathogens can invade and travel through barriers to the distant areas of the body has not been well understood, and is being currently investigated.
P. gingivalis is the most studied oral bacterium due to its ease of growth under laboratory conditions and ability to be genetically manipulated.12 As P. gingivalis reaches the deeper areas of the sulcus/pocket where the environment is favorable for this secondary colonizer, it relies on other species to reduce oxygen concentration in the environment. Considered nonmotile, it has been shown to move under very specific environmental conditions, whether locally or to distant destinations, using what’s known as “gliding motility.”6,13
Found in more than 85% of biofilm samples from periodontal pockets in patients with chronic periodontitis,2 P. gingivalis almost exclusively resides in the three distinct microenvironments of the subgingival crevice: root surface, gingival crevicular fluid, and surface of the gingival epithelial cells lining the crevice. However, it is also present in the tonsillar area, tongue, and buccal mucosa in patients with and without periodontal diseases. It is asaccharolytic (does not rely on digesting sugars for energy production), but is able to ferment amino acids by breaking down the connective tissue proteins. P. gingivalis also requires iron for survival.
Like many other bacterial species, P. gingivalis gets iron from the hemoglobin of the red blood cells, overcoming the host strategies of reducing free iron during infection.14 It is the heme accumulation in the growing P. gingivalis colonies that gives them the unique dark purple-black color, which explains the bacterium’s name: Porphyromonas means “purple units” in Greek.2 Interestingly, P. gingivalis obtains iron using specific outer membrane receptors, proteases known as gingipains and lipoproteins. The bacterium is also capable of evading the host immune response without inflammatory response suppression. Actually, P. gingivalis and other pathogenic bacteria may take advantage of inflammation -deriving nutrients and iron from the inflammatory exudate.6 Twenty-two strains of P. gingivalis with varying degrees of invasiveness and virulence have been identified.15 The genome of one of its most virulent strains—W83—was sequenced in 2003, providing important information on the bacterium’s comparative genomics, metabolism, transport, and its virulence factors.1
Features and Adaptive Abilities Within the Microbiome
Despite the presence of more than 500 bacterial species in human subgingival biofilm, extensive study has shown that P. gingivalis is the predominant etiologic bacterium contributing to chronic periodontitis.2,16 P. gingivalis is rightly termed the “keystone pathogen” because of its ability to use various deliberate mechanisms to resist, weaken, or manipulate host immune responses.9,13,17 P. gingivalis depends on the presence of other biofilm bacteria and the bacterial dysbiosis it initiates. A groundbreaking study in germ-free mice inoculated with P. gingivalis demonstrated its inability to cause periodontitis alone. This research also highlighted the requirement of the intact complement system as part of host immune response in the development of P. gingivalis-induced disease. Mice without functioning C3a/C5a receptors were resistant to developing periodontitis, emphasizing the critical role of these complement signaling pathways and their required manipulation by P. gingivalis in periodontal diseases.7,10
Considering its many adaptive strategies to maintain virulence, P. gingivalis is truly a microbe enigma, much like the bacterium Helicobacter pylori found in the stomach and small intestine. Among its abilities to modify the microbiome’s environment are altering pH imbalances, rectifying oxygen stress, optimizing temperatures, and overcoming countless antagonistic antibacterial peptides and bacteriophages. Recent research into the genome sequencing of P. gingivalis revealed its use of various amino-acid catabolic pathways as an action to resist acidic pH stress in the microbiome. Following the digestion of amino acids, P. gingivalis generates ammonia as a byproduct, which can alter the microbiome from an acidic to neutral pH—ideal for cell proliferation.18
Modifying the functionality of the commensal environment through its virulence factors, P. gingivalis is capable of adapting to a rapidly shifting nutritional environment of “feast or famine” to ensure its nutritional demands for survival.18 As an inflammophilic and asaccharolytic organism, P. gingivalis amasses metabolic and nutritional needs of carbon and nitrogen by digesting peptide accretions from tissue-damaging metabolites, enzymes, and proinflammatory cytokines produced during host inflammation.9,18 P. gingivalis maintains a favorable inflammatory nutrient-rich environment by producing and utilizing its gingipain protease (Arg-gingipain); however, it can transition to using another protease (Lys-gingipain) to ameliorate the negative effects of too high an inflammatory response, highlighting the microbe’s adaptability to its environment.9,16
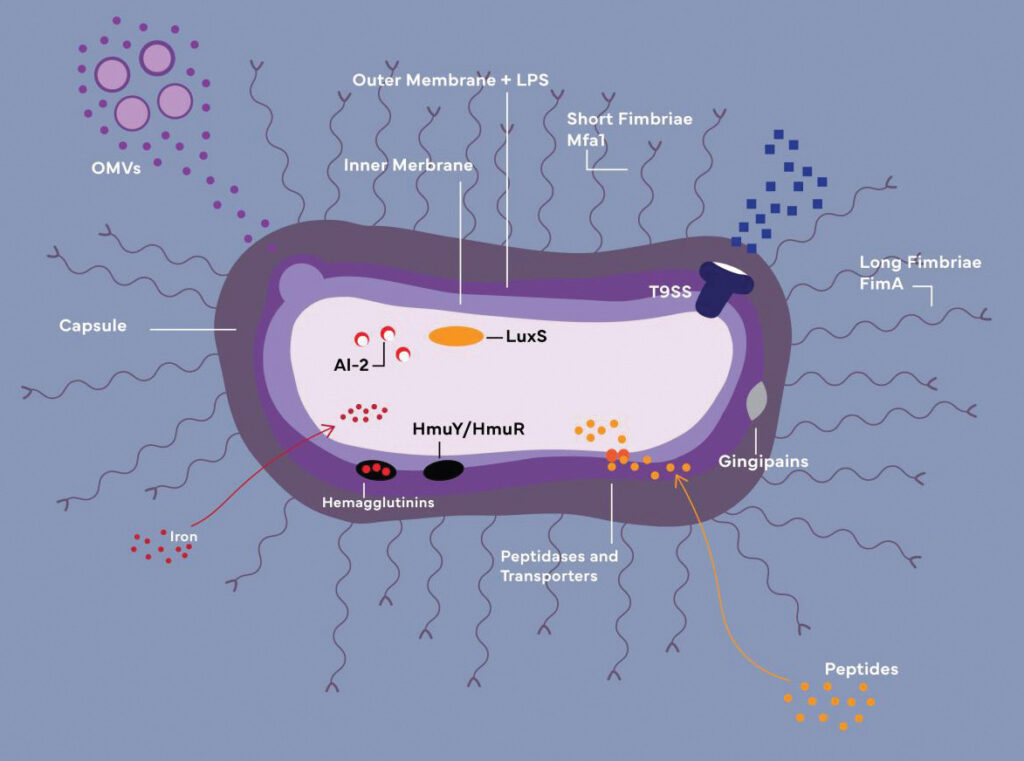
OMVs: Outer membrane vesicles
LPS: Lipopolysaccharide
HmuY/HmuR: Iron-acquiring transporters (function together with
hemagglutinins and gingipains)
AI-2/LuxS: Components of quorum-sensing system
Credit: Cheyenne Syrek
Co-Dependence and Thriving in the Microbiome
Through an intricate organization of unique molecular co-aggregation, P. gingivalis persists within the host by coupling with other microbes and storing portions of the energy-consuming systems discarded by other microbes, subsequently serving the entire bacterial community’s function and growth. P. gingivalis releases gingipains to mediate interactions between complement receptors and to create a nutrient-rich but oxygen-deficient environment as a protective safeguard from the host immune response, thus permitting co-microbe unions to exist.9,10,19 The co-infectious microbes bond by exchanging gene-coding signals using quorum sensing—a bacterial communication mechanism that involves cleaving of specific cellular signaling molecules.19 As P. gingivalis co-aggregates with complement species like T. denticola and T. forsythia, toxic inflammatory outcomes are averted as they breakdown and immobilize numerous antimicrobial complexes and enzymes discharged by neutrophils.9
The latest research on P. gingivalis co-aggregation with Streptococcus gordonii and Filifactor alocis has garnered much interest because of their co-virulence capabilities. Through a complex adhesion system of short and long fimbria, these specific co-aggregates can influence the overall genotype profile of the microbiome and immune response.9,19 F. alocis, a Gram-positive obligate anaerobe is exceptionally resistant to oxidative stress and can vastly modify the microbiome with its own potent virulence factors.20 The union between P. gingivalis and F. alocis form a strong pathobiont bond increasing proliferation of the junctional epithelium and infiltration of collagen fibers in the gingival epithelial cells.9 Similarly, S. gordonii a well-known opportunistic pathogen, strengthens P. gingivalis virulence with its capacity to permeate the pulp tissue and enter the circulatory system.21 The synergistic action between P. gingivalis and S. gordonii destabilizes the immune response in favor of biofilm growth and intensifies oxidative stress, thus sustaining periodontal inflammatory responses beneficial for the survival and proliferation of P. gingivalis.9,10
Perhaps one of the most intriguing abilities of P. gingivalis is its role as both inhibitor and facilitator of cell death. On one hand, P. gingivalis can influence chronic inflammation by precluding programmed cell death (apoptosis), which can be described as controlled cell death without lysis. This prolongs inflammation by longer host cell survival, followed by their lytic death and release of harmful cellular components into the environment, further exacerbating inflammatory response and providing cellular and DNA fragments as nutrients for P. gingivalis and its pathobionts. Conversely, this bacterium can induce suicide of the host’s and other bacterial cells, termed “fratricide,” once again taking advantage of the released cell fragments as nutrients and further facilitating dysbiosis.9
Before P. gingivalis can survive and populate within a host, it must penetrate the host’s exterior protective barriers and transmute the microbiome to a beneficial state. P. gingivalis accomplishes this mission with the assistance of its very potent virulence factors: capsule, outer membrane proteins and vesicles, lipopolysaccharide (LPS), hemagglutinins, fimbriae, gingipains, and others (Figure 1, see page 35).6,16,22 P. gingivalis’ virulence factors under active current investigation are reviewed below.
Gingipains
Ongoing research into the underlying mechanisms of P. gingivalis’ pathogenicity focuses on gingipains—cysteine proteinases anchored to its cell surface.23 Gingipains work by splitting the protein genetic sequences from lysine or arginine residues: lysine-gingipain, arginine-gingipain A, and arginine-gingipain B.2,9,18 These principal gingipains of P. gingivalis are either released into the milieu or become attached to other cell surfaces.23,24 As virulence factors, their role in periodontal tissue degradation and the breakdown of iron-binding proteins is to attack critical components of the extracellular matrix, disrupting the epithelium’s barrier function and allowing P. gingivalis to enter subepithelial tissues.23
Gingipains can also enmesh the circulating proteins necessary for binding apoptotic and necrotic cells and cellular DNA to clean up after injury. These gingipains-proteins interactions interrupt the host’s immunological response, allowing P. gingivalis to remain in the host longer. P. gingivalis can delay the recruitment of neutrophils—an essential step in immune response and a feature in healthy periodontium, which permits the bacterial community time to colonize.9,10 Moreover, by activating and exploiting the C5a-receptor signaling, as mentioned previously, P. gingivalis can block the complement cascade as well as undermine the leukocyte killing capacity.7,9,10
Fimbriae
Fimbriae are pilus appendages generated by gingipains, anchored to the outer cell membrane and required for biofilm production and attacking/infecting target cells while evading the host defense system.19,23 Fimbriae can attach to eukaryotic cells and other bacterial species, allowing for increased motility, biofilm development, and cell penetration.16 Gingipains produce both short and long fimbriae; both types contribute to the production of poly-species biofilms by way of their adhesive capabilities. Long fimbriae not only permit P. gingivalis to adhere to host tissues, but also attach to and suppress the toll-like receptor-mediated pro-inflammatory response, making it possible for P. gingivalis to enter host cells.22,23 The long fimbriae also facilitate the coaggregation of P. gingivalis with other oral pathogens. Also worth highlighting are the functions of Candida albicans, a common oral cavity inhabitant, whose presence inside the polymicrobial biofilm depletes oxygen, shielding P. gingivalis from high-oxygen conditions.23
As a nonmotile bacterium,2,6 P. gingivalis’ capacity to translocate to distant organs has been poorly understood. However, a recent in vitro study discovered promising findings about the motility capabilities of P. gingivalis. The bacterium’s less virulent but more fimbriated strain 381 exhibited a multimodal motility pattern throughout the life cycle under carefully arranged conditions. Throughout the testing, strain 381 bacteria demonstrated motility by rolling over adjacent cells. Remarkably, the capacity of P. gingivalis to roll and displace over the surface of red blood cells was also detected. P. gingivalis was also noted to move by ingesting several diverse metabolites while scavenging and spreading. It appears that rather than being actively motile, P. gingivalis exhibits “gliding motility,“ using surrounding microbes to generate movement through proteolysis, cell dispersal, cell-on-cell rolling, and subdiffusive cell-driven motility, demonstrating how it can translocate to local and distant sites.13
While the experiment demonstrated the significance of one of the proteins produced by P. gingavalis (Mfa5) in coordinating cell motions and cell-on-cell rolling, the exact process is still not fully understood. The T9SS system also plays a vital role in correcting the environment and surface translocation because it releases hygroscopic amino acids that initiate the hydration stage for cell-on-cell rolling and subdiffusive cell-driven motility.13
TS99 Secretion System
The secretion system of a cell plays a significant role in its ability to proliferate within a particular ecological niche by enabling nutrition storage, communicating between cells, defending against antagonists, and inducing virulence factors.25 The first Gram-negative bacterium’s secretion system was discovered in the 1970s, revealing E. coli’s ability to produce hemolysin-A and secrete it into the environment. Since then, eight more secretion pathways have been discovered and are referred to as type x secretion systems (T1SS to T9SS). P. gingivalis’ secretion system (T9SS)—most recently discovered—uses a two-step process, during which the substrates first pass the inner membrane into the periplasm and then are guided to the outer membrane translocation channel.23,25 The T9SS has two different roles: one that is related to bacterial gliding motility, but also a sinister one used by the pathogens fostering dysbiosis. In the latter case, in a pathogenic bacterium such as P. gingivalis, the protein secretions can remain attached to the surface of the outer membrane, released into the extracellular environment, or penetrate the cytoplasm of a target cell.25
Additional Mechanisms
P. gingivalis’ outer membrane is composed of lipopolysaccharide (LPS)—a vital structure necessary to maintain its cell shape and ability to carry out cell functions. LPS functions as a barrier, blocking and preventing assaults or elimination by the host complement. The outer membrane LPS houses enclosed, endocytic-derived outer membrane vesicles that contribute to toxin transport and pathogenicity.4,23 Recent findings demonstrate that bacterial pathogens like P. gingivalis activate outer membrane vesicles as a means of communication, manipulation, and disruption of the normal immune response of the host, revealing once again the profound ingenuity and capabilities of this microorganism.18
Peptidylarginine deiminase (PPAD) is a protein-modifying enzyme converting arginine residues to neutral citrulline residues, altering protein structure and function. This process, known as citrullination, is a major factor influencing colonization and biofilm growth. Thus far, P. gingivalis is the only bacterium in the human body known to generate and secrete PPAD,11,23 and its precise role in periodontal diseases and bacterial biochemistry is yet unknown, studies suggest that ammonia—a byproduct of PPAD activity—aids P. gingivalis’ resistance to elimination through acidic cleansing, a process initiated by the host immune response.13 This view is supported by the restriction of P. gingivalis’ growth on protein substrates at low pH, and observation of citrullination and amino acid fermentation of lysine and arginine providing a favorable environment for its survival. PPAD can be secreted with other virulence factors (gingipains) directly through the T9SS or indirectly through outer membrane vesicles.11,17,23
Conclusion
P. gingivalis is an especially dangerous and capable periodontal pathogen, affecting not only oral but also systemic health. This review of P. gingavalis and its virulence factors is far from complete as ongoing research reveals more insights into how the bacterium lives and succeeds as one of the most notable periodontal pathogens.
References
- Nelson KE, Fleischmann RD, DeBoy RT, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–5601.
- How KY, Song KP, Chan KG. Porphyromonas gingivalis: an overview of periodontopathic pathogen below the gum line. Front Microbiol. 2016;7:53.
- Cutler CW, Kalmar JR, Genco CA. Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 1995;3:45–51.
- Zhang Z, Liu D, Liu S, Zhang S, Pan Y. The role of Porphyromonas gingivalis outer membrane vesicles in periodontal disease and related systemic diseases. Front Cell Infect Microbiol. 2021;10.
- Viafara-García SM, Morantes SJ, Chacon-Quintero Y, Castillo DM, Lafaurie GI, Buitrago DM. Repeated Porphyromonas gingivalis W83 exposure leads to release pro-inflammatory cytokynes and angiotensin II in coronary artery endothelial cells. Sci Rep. 2019;9:19379.
- Mysak J, Podzimek S, Sommerova P, et al. Porphyromonas gingivalis: major periodontopathic pathogen overview. J Immunol Res. 2014;2014:e476068.
- Hajishengallis G, Liang S, Payne MA, et al. A low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and the complement pathway. Cell Host Microbe. 2011;10:497–506.
- Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30.
- Chopra A, Bhat SG, Sivaraman K. Porphyromonas gingivalis adopts intricate and unique molecular mechanisms to survive and persist within the host: a critical update. J Oral Microbiol. 2020;12:1801090.
- Hajishengallis G, Lamont RJ. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol. 2014;44:328–338.
- Vermilyea DM, Ottenberg GK, Davey ME. Citrullination mediated by PPAD constrains biofilm formation in P. gingivalis strain 381. NpJ Biofilms Microbiomes. 2019;5:1–11.
- Chen T, Hosogi Y, Nishikawa K, et al. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J Bacteriol. 2004;186:5473–5479.
- Moradali MF, Ghods S, Angelini TE, Davey ME. Amino acids as wetting agents: surface translocation by Porphyromonas gingivalis. ISME J. 2019;13:1560–1574.
- Richard KL, Kelley BR, Johnson JG. Heme uptake and utilization by Gram-negative bacterial pathogens. Front Cell Infect Microbiol. 2019;9:81.
- Igboin CO, Griffen AL, Leys EJ. Porphyromonas gingivalis strain diversity. J Clin Microbiol. 2009;47:3073–3081.
- Mei F, Xie M, Huang X, et al. Porphyromonas gingivalis and its systemic impact: current status. pathogens. 2020;9:944.
- Bereta G, Goulas T, MadeJ M, et al. Structure, function, and inhibition of a genomic/clinical variant of Porphyromonas gingivalis peptidylarginine deiminase. Protein Sci. 2019;28:478-486.
- Nara PL, Sindelar D, Penn MS, Potempa J, Griffin WST. Porphyromonas gingivalis outer membrane vesicles as the major driver of and explanation for neuropathogenesis, the cholinergic hypothesis, iron dyshomeostasis, and salivary lactoferrin in Alzheimer’s disease. J Alzheimers Dis JAD. 2021;82:1417–1450.
- Gerits E, Verstraeten N, Michiels J. New approaches to combat Porphyromonas gingivalis biofilms. J Oral Microbiol. 2017;9:1300366.
- Aja E, Mangar M, Fletcher HM, Mishra A. Filifactor alocis: recent insights and advances. J Dent Res. 2021;100:790–797.
- Park OJ, Kwon Y, Park C, et al. Streptococcus gordonii: pathogenesis and host response to its cell wall components. microorganisms. 2020;8:1852.
- Jia L, Han N, Du J, Guo L, Luo Z, Liu Y. Pathogenesis of important virulence factors of Porphyromonas gingivalis via toll-like receptors. Front Cell Infect Microbiol. 2019;9:262.
- Lunar Silva I, Cascales E. Molecular strategies underlying Porphyromonas gingivalis virulence. J Mol Biol. 2021;433:166836.
- Patel S, Howard D, French L. Susceptibility to gingipains and transcriptomic response to P. gingivalis highlights the ribosome, hypothalamus, and cholinergic neurons. Available at: biorxiv.org/content/葖.1101/떔.08.09.243402v1. Accessed September 22, 2022.
- Lasica AM, Ksiazek M, MadeJ M, Potempa J. The type ix secretion system (t9ss): highlights and recent insights into its structure and function. Front Cell Infect Microbiol. 2017;7:215.
From Dimensions of Dental Hygiene. October 2022; 20(10)34-37.



