 GOPIXA/ISTOCK/GETTY IMAGES PLUS
GOPIXA/ISTOCK/GETTY IMAGES PLUS
The Role of Genetics in Caries Risk and Resistance
Improved understanding of relevant genetic factors will help increase the precision of caries risk assessment and, ultimately, enable more targeted, personalized interventions that will effectively prevent and manage dental caries.
This course was published in the February 2019 issue and expires February 2022. The author has no commercial conflicts of interest to disclose. This 2 credit hour self-study activity is electronically mediated.
EDUCATIONAL OBJECTIVES
After reading this course, the participant should be able to:
- Discuss the role genetics plays in an individual’s risk or resistance to dental caries.
- Explain common concepts and curricula that shape clinician’s outlook on the caries process.
- Describe the microbiome and its relationship to caries and health, and the importance of a diverse oral microbiota.
Although caries risk assessments evaluate an individual’s caries experience, they tend not to assess the individual’s family history of caries to determine if heritability is contributing to the disease.5,6 The concept that genetics play a role is certainly not new, as many animal and human studies have shown the importance of genetics as a risk factor.7,8 Studies of twins have been used for more than 50 years to evaluate the potential role of heredity in caries.9,10 These studies routinely show a higher concordance of caries in monozygotic twins compared with dizygotic twins or siblings, supporting the role of genetics as a caries determinant.11–13
Not surprisingly, there are many genetic factors that likely contribute to caries risk and resistance, such as taste preference, salivary factors, immune response, tooth morphology, enamel composition and structure, and behavior. Dental caries is a chronic complex disease, so it is likely that multiple genes contribute to caries risk/ resistance vs a gene of major effect, such as occurs in Mendelian inheritance. There also can be gene-gene interactions or gene-environment interactions creating epigenetic effects that all possibly contribute to risk and resistance. This manuscript presents science’s current understanding of the genetic risk and resistance determinants for caries, and how this knowledge could be used in disease management.
GENETIC CARIES DETERMINANTS RELATED TO DIET AND TASTE
The relationship between caries, diet, and genetics is an excellent example of the nature/ nurture interplay affecting a disease process. Hereditary conditions that cause dietary modifications are associated with either decreased and increased caries experience. Hereditary fructose intolerance is a rare autosomal recessive hereditary condition caused by mutations in the gene coding for aldose B. Due to the inability to effectively metabolize fructose and sucrose, affected individuals markedly restrict their intake—resulting in fewer caries lesions compared with unaffected individuals.14 Patients with generalized recessive dystrophic epidermolysis bullosa have marked mucosal oral fragility and scarring, along with the need for high caloric intake to support wound healing and compensate for poor nutrient uptake.15 Affected individuals typically eat slowly, have delayed oral clearance of food, and consume carbohydrate-rich diets; collectively, these result in a high caries rate.
Taste preference is determined by genetics and environmental exposures. The link between taste preference and dental caries is not a new concept.16 The human sense of taste evolved to help detect important sources of nutrients (eg, sugars, salts, and amino acids) while protecting the individual from poisonous substances (eg, acids and alkaloids).17 Taste buds and taste receptors reside in the oral cavity largely on the tongue, and are coded by families of genes that reside in clusters in the human genome.17 Studies suggest associations of differing caries levels in people with genetic variations in their taste genes.18,19 The TAS1R1 and TASIR3 genes code for a taste receptor that mediates umami (distinct form of saltiness) perception. The TAS1R1 gene is located on chromosome 1p36 and is associated with a cluster of other taste receptor genes, such as TAS1R2 that codes for a receptor involved in the perception of sweetness. The TASR38 taste receptor gene that is involved in perception of glucosinolates has been associated with lower caries rates.18 Other genes that may be involved in dietary preference that are associated with caries include the guanine nucleotide binding protein, alpha transducing 3 (GNAT3) gene.18 Thought to play a role in transduction of taste perception, gustaducin is the protein product of the GNAT3 gene and is present in all forms of taste buds. Evaluation of caries and sucrose taste preference in twins reveals that both are associated with genetic determinants, and understanding the heritability of these traits may add to a clinician’s ability to assess individual caries risk.20 Table 1 provides an overview of several known caries-determinant genes and their functions.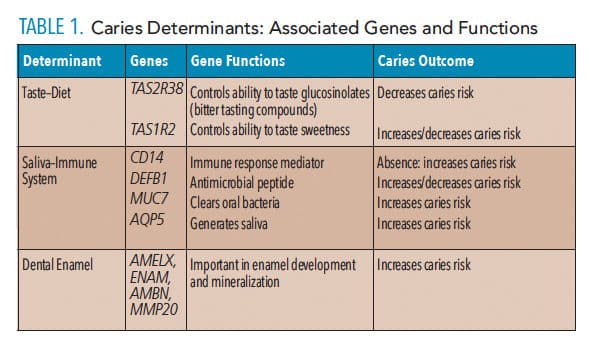
SALIVA AND THE IMMUNE SYSTEM
Saliva serves as a critical defense mechanism to caries by providing an aqueous medium for fluoride, calcium, and phosphate ions for remineralization, buffering, lubrication, digestive capacity of food substrates, and immunological factors, to name just a few essential functions. Hereditary conditions associated with abnormal salivary production, such as aplasia of lacrimal and salivary glands, are associated with increased caries experience. This autosomal-dominant condition is caused by mutations in the FGF10 gene that is critical for normal gland development, resulting in salivary gland aplasia and a drastic reduction in salivary production and flow. Genetic variations in alleles coding for the protein HaeIII subfamily 1 (PRH1) gene are associated with both Streptococcus mutans colonization and caries risk.21 Given the many anticaries attributes of saliva, it is not surprising that multiple salivary genes are associated with caries risk or resistance.22
A certain segment of the population is considered to be immune from dental caries (ie, highly protected and unaffected), and part of this resistance is thought to emanate from the immune system. Saliva provides a rich medium to support the oral cavity’s immune response and does so with many genes that have antimicrobial properties, such as the salivary peroxidase system. Several genes that code for proteins with antimicrobial properties that are secreted in saliva have been associated with caries susceptibility, such as lactotransferrin.23 Variations in the salivary protein T-Cell Receptor Alpha Chain Variable 4, and the gene that codes for this protein (TRAV4), are associated with low caries experience.24 Individuals with a missense mutation in TRAV4 have higher caries rates, and this mutation is predicted to cause a reduction in the protein’s protective potential.24 The antimicrobial protein Beta defensin 1 (DEFB1) is secreted in saliva and is thought to support the defense of the oral epithelia surfaces; it has also been implicated as being protective against both caries and periodontal disease. Haplotype studies of the DEFB1 gene suggest that specific allelic variants are associated with either an increased or decreased decayed/ missing/ filled teeth score. These functional polymorphisms in the DEFB1 gene are possible markers for caries risk or resistance.25 While numerous salivary proteins involved in oral immunity—such as mucins—may be determinants in caries experience, the role of genetics and genetic variability in risk and resistance is yet to be defined.26,27
GENETICS OF TOOTH-ASSOCIATED DETERMINANTS OF CARIES
Dental caries is the process of demineralizing the enamel, dentin, and cementum, and the quality of those tissues can influence initiation and progression of disease. Development of the dentition is the culmination of diverse and complex processes that are orchestrated through strict molecular control. Tooth formation is influenced by environmental factors, including diet, infection, and trauma. Evidence indicates that an individual’s response to environmental exposures, such as fluoride, can vary based on the patient’s genetic constitution.28 Enamel formation involves the expression of thousands of genes, and more than 100 conditions are associated with abnormal enamel development.29
Numerous environmental exposures and stressors are also associated with developmental defects in enamel. Consequently, such defects are highly prevalent. Enamel defects are associated with early and greater colonization with streptococcus mutations and the formation of lesions.30 It has been proposed that certain cases of early childhood caries are directly due to enamel hypoplasia that results in early development and progression of severe disease.31
Developmental defects that appear to involve genetic-environmental interactions can add to an individual’s risk or resistance to caries. Individuals with X-linked nephrogenic diabetes insipidus often suffer from dental fluorosis due to polydipsia and renal dysfunction.32 First permanent molars frequently have enamel hypomineralization (about 5% to 30% of populations around the world are affected) and are also frequently affected with caries.33 Molar incisor hypomineralization has traditionally been thought to have an environmental etiology, however, more recent studies suggest genetic variants may contribute to an individual’s risk for this condition.34–36
Genes that are critical for normal enamel formation, including amelogenin (AMELX), enamelin (ENAM), matrix metalloproteinase 20 (MMP20) and kallikrein 4 (KLK4), are all associated with hereditary enamel malformations known as amelogenesis imperfecta.37 Variants in the genetic code of these genes that do not cause amelogenesis imperfecta have been associated with either increased or decreased caries experience.38,39
Humans with mutations in genes coding for laminin type V, (ie, LAMA3) can have junctional epidermolysis bullosa that is associated with fragility of the skin and blistering, as well as enamel hypoplasia due to abnormal cell-cell attachment of the ameloblasts.40 Individuals with this autosomal recessive trait with thin, pitted enamel face markedly increased risk for developing caries.41 Although not well understood, the genetic determinants of the pits and fissures of teeth also are likely involved in contributing to the heritability of caries. Studies have mapped pit and fissure caries in families, showing strong familiality of the trait. Other enamel-associated genes, including ameloblastin (AMBN), tuftelin (TUFT1), and tuftelin-interacting protein 11 (TFIP11) also have been associated with variation in caries rates.
ORAL MICROBIOME
For more than a decade, the Human Microbiome Project has been studying the role of microbes in human health and disease. Researchers have characterized the microbial communities inhabiting various body surfaces, including the oral cavity. Over the last two decades, new technologies, such as 16S rRNA gene sequencing, have allowed a more comprehensive understanding of the oral microbiome and its relationship to caries and health. The human oral microbiome is extremely diverse, with more than 600 taxa and likely twice that many microbial species.42,43 Maintaining a stable and healthy oral microbiota associated with oral health—in contrast to dysbiosis, or a microbial shift toward disease, and how this shift occurs—are concepts that are not fully understood.44
Metagenomics, or the genomic research into uncultured microorganisms, allows the study of the diverse microbiota in the oral cavity and how it changes in conditions of health and disease. This discipline is likely to contribute to highly personalized care in dental caries management.44 Knowledge gained using these approaches indicates that caries-associated microorganisms are more diverse than previously believed. In addition, greater microbial diversity is seen in children without caries than in those with the disease.45 Studies of the gut microbiota show their importance in influencing human development, metabolism, immunology, and disease. Advanced molecular biology techniques are helping to reveal the complex relationships between the human genome and microbiome.46
SUMMARY
Scientific understanding of genetics and its role in caries has advanced tremendously over the past 60 years, but remains quite incomplete. Twin studies provide an interesting model for determining the nature-vs-nurture aspects of caries, as most studies show strong genetic contributions to caries experience. Given the complex nature of dental caries, with the many diverse contributing determinants of disease and genetic/ environmental interactions, it is understandable that numerous genes are being implicated in caries risk and resistance. The goal of studies to determine potentially important genetic factors is to allow more precise caries risk assessment, and, ultimately, more targeted, personalized interventions that will effectively prevent and manage the disease.
REFERENCES
- Petersen PE, Bourgeois D, Ogawa H, Estupinan-Day S, Ndiaye C. The global burden of oral disease and risks to oral health. Bull World Health Organ. 2005;83:661–669.
- Tanzer JM. Dental caries is a transmissible infectious disease: the Keyes and Fitzgerald revolution. J Dent Res. 1995;74:1536–1542.
- Caufield PW. Dental caries: an infectious and transmissible disease where have we been and where are we going? N Y State Dent J. 2005;71:23–27.
- Brown JP. A new curriculum framework for clinical prevention and population health, with a review of clinical caries prevention teaching in U.S. and Canadian dental schools. J Dent Educ. 2007;71:572–578.
- Pitts N, Melo P, Martignon S, Ekstrand K, Ismail A. Caries risk assessment, diagnosis and synthesis in the context of a European Core Curriculum in Cariology. Eur J Dent Educ. 2011;15 Suppl 1:23–31.
- Doméjean S, White JM, Featherstone JD. Validation of the CDA CAMBRA caries risk assessment — a six-year retrospective study. J Calif Dent Assoc. 2011;39:709–715.
- Kurihara Y, Naito T, Obayashi K, Hirasawa M, Kurihara Y, Moriwaki K. Caries susceptibility in inbred mouse strains and inheritance patterns in F1 and backcross (N2) progeny from strains with high and low caries susceptibility. Caries Res. 1991;25:341–346.
- Vieira AR, Modesto A, Marazita ML. Caries: review of human genetics research. Caries Res. 2014;48:491–506.
- Horowitz SL, Osborne RH, Degeorge FV. Caries experience in twins. Science. 1958;128:300–301.
- Finn SB, Caldwell RC. Dental caries in twins – I. A comparison of the caries experience of monozygotic twins, dizygotic twins and unrelated children. Arch Oral Biol. 1963;8:571–585.
- Boraas JC, Messer LB, Till MJ. A genetic contribution to dental caries, occlusion, and morphology as demonstrated by twins reared apart. J Dent Res. 1988;67:1150–1155.
- Conry JP, Messer LB, Boraas JC, Aeppli DP, Bouchard TJ Jr. Dental caries and treatment characteristics in human twins reared apart. Arch Oral Biol. 1993;38:937–943.
- Bretz WA, Corby PM, Hart TC, et al. Dental caries and microbial acid production in twins. Caries Res. 2005;39:168–172.
- Newbrun E, Hoover C, Mettraux G, Graf H. Comparison of dietary habits and dental health of subjects with hereditary fructose intolerance and control subjects. J Am Dent Assoc. 1980;101:619–626.
- Haynes L. Nutrition for children with epidermolysis bullosa. Dermatol Clin. 2010;28:289–301.
- Chung CS, Witkop CJ, Henry JL. A genetic study of dental caries with special reference to PTC taste sensitivity. Am J Hum Genet. 1964;16:231–245.
- Kinnamon SC. A plethora of taste receptors. Neuron. 2000;25:507–510.
- Wendell S, Wang X, Brown M, et al. Taste genes associated with dental caries. J Dent Res. 2010;89:1198–1202.
- Kulkarni GV, Chng T, Eny KM, Nielsen D, Wessman C, El-Sohemy A. Association of GLUT2 and TAS1R2 genotypes with risk for dental caries. Caries Res. 2013;47:219–225.
- Bretz WA, Corby PM, Melo MR, et al. Heritability estimates for dental caries and sucrose sweetness preference. Arch Oral Biol. 2006;51:1156–1160.
- Zakhary GM, Clark RM, Bidichandani SI, Owen WL, Slayton RL, Levine M. Acidic proline-rich protein Db and caries in young children. J Dent Res. 2007;86:1176–1180.
- Wang X, Shaffer JR, Zeng Z, et al. Genome-wide association scan of dental caries in the permanent dentition. BMC Oral Health. 2012;12:57.
- Azevedo LF, Pecharki GD, Brancher JA, et al. Analysis of the association between lactotransferrin (LTF) gene polymorphism and dental caries. J Appl Oral Sci. 2010;18:166–170.
- Briseño-Ruiz J, Shimizu T, Deeley K, et al. Role of TRAV locus in low caries experience. Hum Genet. 2013;132:1015–1025.
- Ozturk A, Famili P, Vieira AR. The antimicrobial peptide DEFB1 is associated with caries. J Dent Res. 2010;89:631–636.
- Pol J. [Association of the polymorphism of MUC7 gene encoding the low-molecular-weight mucin MG2 with susceptibility to caries]. Ann Acad Med Stetin. 2011;57:85–91.
- Gabryel-Porowska H, Gornowicz A, Bielawska A, et al. Mucin levels in saliva of adolescents with dental caries. Med Sci Monit. 2014;20:72–77.
- Everett ET, Yin Z, Yan D, Zou F. Fine mapping of dental fluorosis quantitative trait loci in mice. Eur J Oral Sci. 2011;119 (Suppl 1):8–12.
- Wright JT, Carrion IA, Morris C. The molecular basis of hereditary enamel defects in humans. J Dent Res. 2015;94:52–61.
- Li Y, Navia J, Caufield P. Colonization by mutans streptococci in the mouths of 3- and 4-year-old Chinese children with or without enamel hypoplasia. Archs Oral Biol. 1994;39:1057–1062.
- Caufield PW, Li Y, Bromage TG. Hypoplasia-associated severe early childhood caries — a proposed definition. J Dent Res. 2012;91:544–550.
- Seow WK, Thomsett MJ. Dental fluorosis as a complication of hereditary diabetes insipidus: studies of six affected patients. Pediatr Dent. 1994;16:128–132.
- Allazzam SM, Alaki SM, El Meligy OA. Molar incisor hypomineralization, prevalence, and etiology. Int J Dent. 2014;2014:234508.
- Jälevik B1, Norén JG. Enamel hypomineralization of permanent first molars: a morphological study and survey of possible aetiological factors. Int J Paediatr Dent. 2000;10:278–289.
- Kühnisch J, Thiering E, Heitmüller D, et al. Genome-wide association study (GWAS) for molar-incisor hypomineralization (MIH). Clin Oral Investig. 2014;18:677–682.
- Jeremias F, Koruyucu M, Küchler EC, et al. Genes expressed in dental enamel development are associated with molar-incisor hypomineralization. Arch Oral Biol. 2013;58:1434–1442.
- Wright JT, Torain M, Long K, et al. Amelogenesis imperfecta: genotype-phenotype studies in 71 families. Cells Tissues Organs. 2011;194:279–283.
- Wang X, Willing MC, Marazita ML, et al. Genetic and environmental factors associated with dental caries in children: the Iowa Fluoride Study. Caries Res. 2012;46:177–184.
- Bayram M, Deeley K, Reis MF, et al. Genetic influences on dental enamel that impact caries differ between the primary and permanent dentitions. Eur J Oral Sci. 2015;123:327–334.
- Wright JT. Oral manifestations in the epidermolysis bullosa spectrum. Dermatol Clin. 2010;28:159–164.
- Wright JT, Fine JD, Johnson L. Dental caries risk in hereditary epidermolysis bullosa. Pediatr Dent. 1994;16:427–432.
- Jenkinson HF. Beyond the oral microbiome. Environ Microbiol. 2011;13:3077–3087.
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010:baq013.
- Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012;18:109–120.
- Ling Z, Kong J, Jia P, et al. Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb Ecol. 2010;60:677–690.
- Wang J, Thingholm LB, Skiecevičienė J, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48:1396–1406.
From Dimensions of Dental Hygiene. February 2019;17(2):26–28,31.



