
Study: Antibody Points to Possible Achilles Heel on Novel Coronavirus
As scientists work tirelessly to develop a vaccine for the novel coronavirus, researchers at The Scripps Research Institute, La Jolla, California, along with colleagues at The University of Hong Kong, are working to produce accurate 3D molecular maps of the virus
As scientists work tirelessly to develop a vaccine for the novel coronavirus, researchers at The Scripps Research Institute, La Jolla, California, along with colleagues at The University of Hong Kong, are working to produce accurate 3D molecular maps of the virus. These molecular maps will guide the development of an effective and safe vaccine to fight the pandemic.
The paper, “A Highly Conserved Cryptic Epitope in the Receptor-Binding Domains of SARS-CoV-2 and SARS-CoV,” published in the journal Science, used high-resolution X-ray crystallography tools to learn how the human system interacts with the virus and kills it. In Wilson’s National Institutes of Health (NIH)-funded lab, the team captured the atomic structure of the antibody bound to its target by shooting X-rays through its crystallized form.
Using the CR3002 antibody from a person who recovered from severe acute respiratory syndrome (SARS), the team investigated where the antibody attaches to the novel coronavirus. Those sites on the virus may be vulnerable to attack, which may inform scientists currently developing vaccines. Previous research has shown that while CR3002 cross-reacts with the novel coronavirus, it does not bind tight enough to neutralize and stop it from infecting cells, according to the NIH.
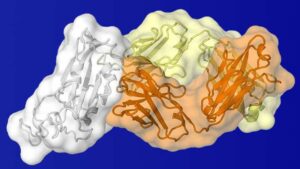
“It is important to find where antibodies are produced against SARS-CoV-2. If these are potent and neutralizing, they can identify sites of vulnerability on the virus that can be used for vaccine and therapeutic design. They also indicate whether infected individuals are likely to be protected from subsequent infection, but it is too soon to tell for how long,” says Ian Wilson, DPhil, Hansen Professor of Structural Biology, and chairman of the Department of Integrative Structural and Computational Biology California Campus.
The new paper found that the CR3002 antibody binds to a similar site on both the SARS and novel coronaviruses. By binding to a spike protein, the antibody is able to bind to the receptor proteins on the surface of the human cells, ACE2. This act of bInding to those receptor proteins is how these viruses first gain entry to human cells and infect them. Researchers found the antibody binds onto the virus’s spike protein at a different location than where the human ACE2 protein binds to the novel coronavirus.
The study uncovered one potential vulnerability of the novel coronavirus: a spot on the virus’s spike protein that is usually hidden except for when the virus shapeshifts its structure in order to infect a cell.
“This antibody bound to a buried site on the receptor binding domain of the SARS-2 spike protein. This binding site is likely transiently exposed but more fully available when the receptor binding domain (RBD) is in the up conformation in which the ACE2 receptor is bound,” Wilson explains. The receptor binding site is also hidden on the spike protein when the RBD is in the down conformation.
This finding suggests an effective vaccine may one that elicits antibodies that target this spot but binds more tightly.
Wilson and his team are currently awaiting neutralizing antibodies isolated from convalescent patients by their collaborators at Scripps Research and elsewhere so they can then map these vulnerable sites on the SARS-CoV-2 spike protein.

